EGA Webin
EGA Webin serves as a platform for registering metadata for array based submissions, large scale sequence submission as well as for legacy EGA submission accounts (ega-box-XXXX). For large scale submitters of sequence data you have also the option to submit metadata via XMLS programmatic submission, while new submitters are advised to utilise the Submitter Portal for their submissions.
WEBIN actions:
-
Register metadata for a sequence submission
- Register study, samples, experiments, runs, DAC, policy and dataset/s after file upload.
-
Register components for your array-based metadata submission
- Register study, samples, DAC or policy before uploading files.
-
Edit existing submission metadata
- Change or update previously submitted metadata.
Register metadata for a sequence submission
Ensure that all sequence files have been encrypted before uploading them to your submission account using the EgaCryptor.
Go to the EGA Webin and log in using your submission account name (ega-box-XXX) and password.
Register components of your metadata submission
- Study
- Samples
- Data Access Committee (DAC)
- Data access policy
- Dataset
For array-base submissions: Study, Samples, Data Access Committee (DAC) and Data access policy may all be registered BEFORE file upload and dataset registration through the array-base template.
Register your Study
- Go to the “Studies (Projects)” box
- Click on “Register Study” and fill in the information related to your study.
- Click on “Submit”, this will save the information and generate an EGA study ID.
To use the study accession number in a publication, the study has to be previously released on the EGA website, we suggest the following format:
"Sequence data has been deposited at the European Genome-phenome Archive
(EGA), which is hosted by the EBI and the CRG, under accession number
EGASXXXXXXXXXXX.
Further information about EGA can be found on
https://ega-archive.org and “The European Genome-phenome Archive in 202 "(10.1093/nar/gkab1059)"
Register your Samples
- Go to the “Samples” box
-
Click on “Register Samples”
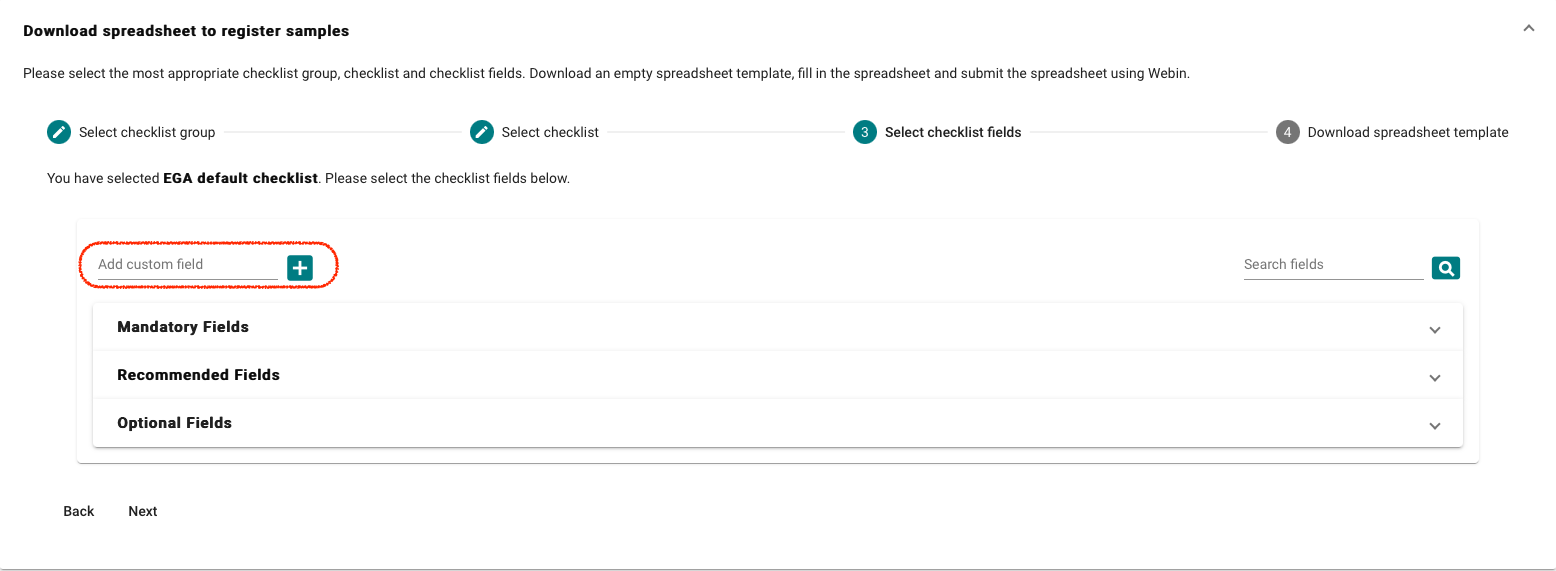
- Select “Download spreadsheet to register samples” and customise your template, there is a default EGA template (EGA default checklist) but more attributes can be added if required.
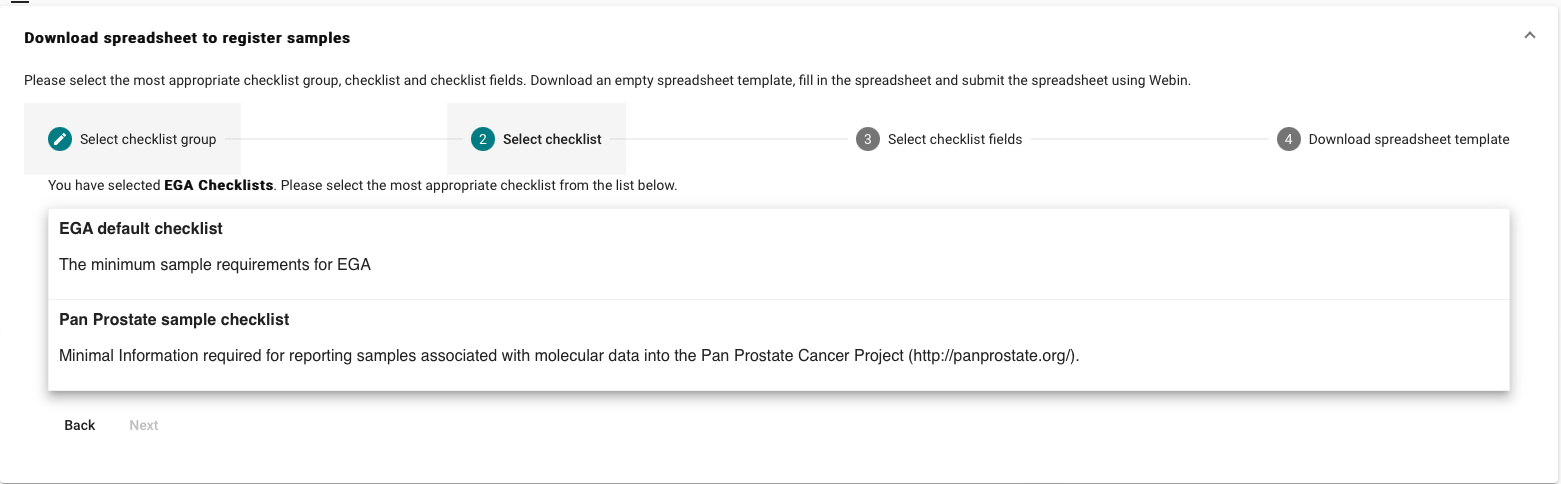
For the EGA default checklist, there are mandatory,recommended and optional
attributes. As well custom fields which can be added if required.
Mandatory attributes
| Field Name | Description |
| tax_id | Taxonomy ID of the organism as in the NCBI Taxonomy database. Entries in the NCBI Taxonomy database have integer taxon IDs. See our tips for sample taxonomy here |
| scientific_name | Scientific name of the organism as in the NCBI Taxonomy database. Scientific names typically follow the binomial nomenclature. For example, the scientific name for humans is Homo sapiens. |
| sample_alias | Unique name of the sample. If not selected system will auto generate an unique alias |
| sample_title | Title of the sample |
| sample_description | Description of the sample |
| phenotype *** | Where possible, please use the Experimental Factor Ontology (EFO) to describe your phenotypes. |
Recommended attributes
| Field Name | Description |
| subject_id | Identifier for the subject where the sample has been derived from |
| gender * | Sex |
Optional attributes
| Field Name | Description |
| sex | sex of the organism from which the sample was obtained |
| disease_site |
Affected organ |
| sample type |
Affected organ |
| donor_id ** |
Identifier of the donor where the sample has been derived from |
*Gender should be described as 'male', 'female' or 'unknown'. If 'unknown' due to a known sex chromosome aneuploidy, please create a user defined attribute called 'Sex chromosome karyotype' and add the appropriate value, for example, 'XXY'.
**Donor id (Subject id) should be a de-identified subject handle. If unknown, please add 'unknown' to the field.
***Phenotypes should, where possible, be an Experimental Factor Ontology accession. If a term cannot be found to describe your phenotype please use free text. All sample phenotypes considered important for further analysis of the data should be provided (for example, tumour type), additional phenotype attributes can be created by defining your own attributes; use the notion 'phenotype2', 'phenotype3', etc.
- After you have customised the fields for the sample submission, download the template and fill in the information.
- Example of the sample template:
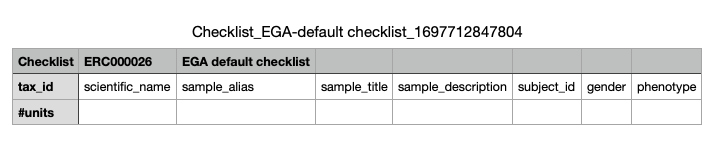
- Finally upload the sample template to get the EGA accession IDs for the samples.

Register your Data Access Committee (DAC)
Further information on the role of your DAC
- Go to the “Data Access” box
- Click on “Register Dacs”
- Input the information about the DAC and register at least one main DAC contact.
Register your Data Access Policy
Your Data Access Policy provides the terms and conditions of data use, this is also referred to as the Data Access Agreement (DAA). Completion of a DAA by the applicant/s should form part of the application process to the Data Access Committee (DAC).
- Go to the “Data Access” box
- Click on “Register Policies”
- Select the DAC to which this policy will be linked to and fill in the policy information.
Submitting your Runs and Analyses
This section is only for sequence data submission, for array-based submission it can be skipped. Please refer to our Submitting array based metadata
Runs Registration
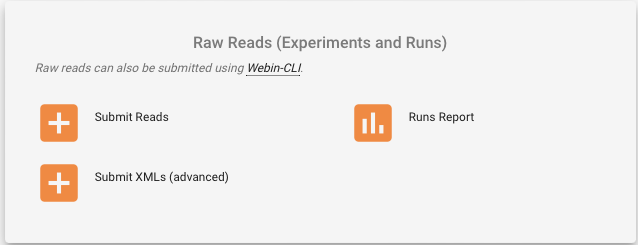
- Go to the “Raw Reads (Experiments and Runs)” box
- Click on “Submit Reads”
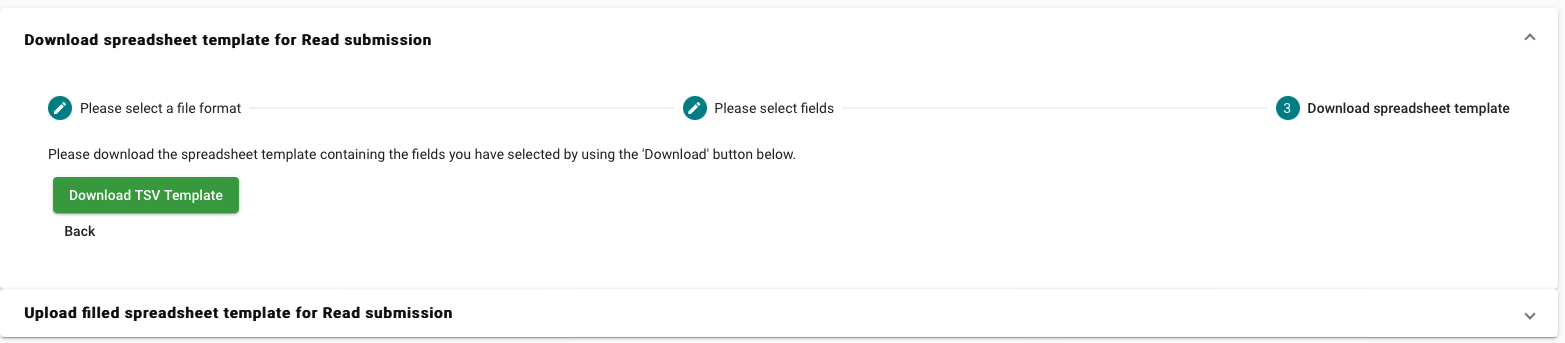
- Select “Download spreadsheet template for Read submission”
- Select the template corresponding to your submission type
- For the templates you have the option to customise the optional fields. To check their description click on “Show Description “
-
Download the template and fill in the required information.
-
Example of the runs template:
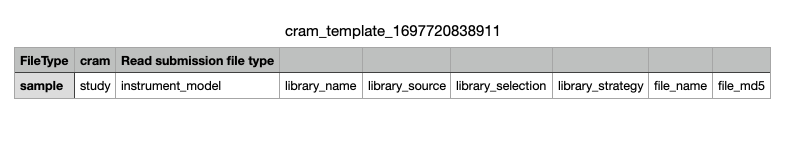
-
Example of the runs template:
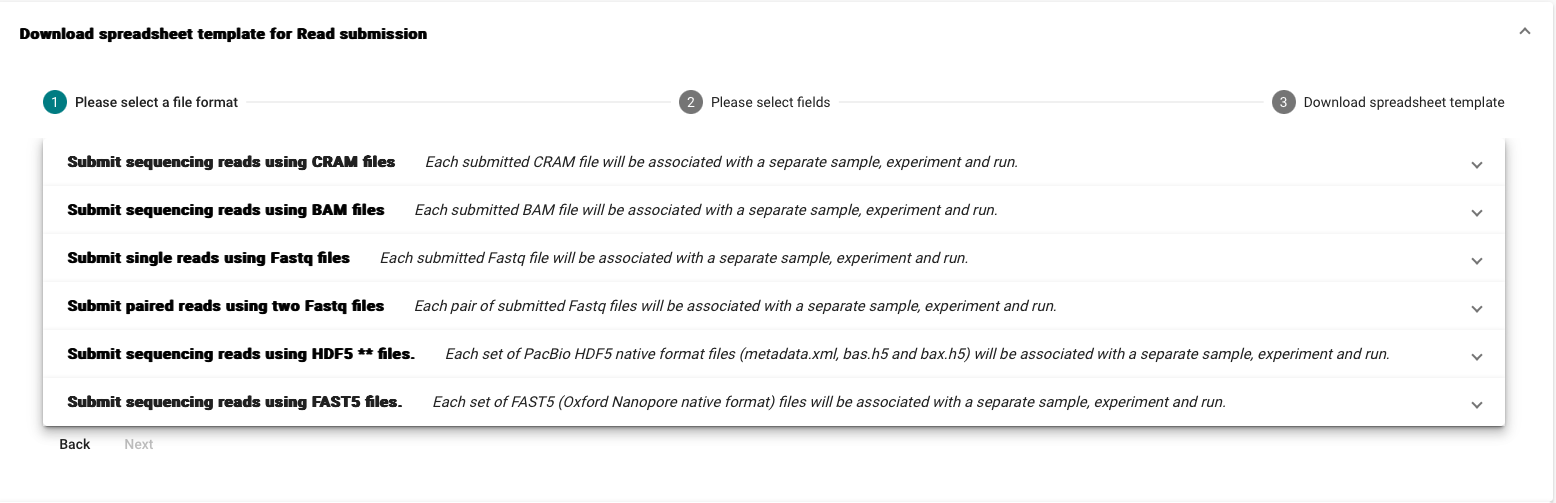
We recommend that Fastq, BAM, and CRAM read files are submitted using Webin-CLI
When using this interface instead of Webin-CLI, raw sequences must be uploaded in one of the supported data formats before they can be submitted. The files can be uploaded using FTP or Aspera.
The study and the sequenced samples must be pre-registered before the raw reads are submitted. Please note that each individual study and sample should be registered only once. You will be asked to provide information about the sequencing libraries and instruments.
Submitting your Dataset
This section is only for sequence data submissions, for array based submissions it can be skipped. Please refer to our Submitting array based metadata
The dataset describes the data files, defined by the run (EGARXXXXXXXXXXX) and analysis (EGAZXXXXXXXXXXX) accessions that make up the dataset and links the collection of data files to a specified Data Access Committee and Data Access Policy.
As a result, you must have registered your Reads and experiments, Data Access Committee (DAC) and Data access policy before submitting your Dataset.
Please consider the number of datasets that your submission consists of, for example, a case control study is likely to consist of at least two datasets. In addition, we suggest that multiple datasets should be described for studies using the same samples but different sequence technologies. Please contact EGA Helpdesk for further assistance.
- Go to the “Data Access” box
- Click on “Register Dataset”
- Select the Data Access Committee (DAC) and Data access policy
-
Register your dataset
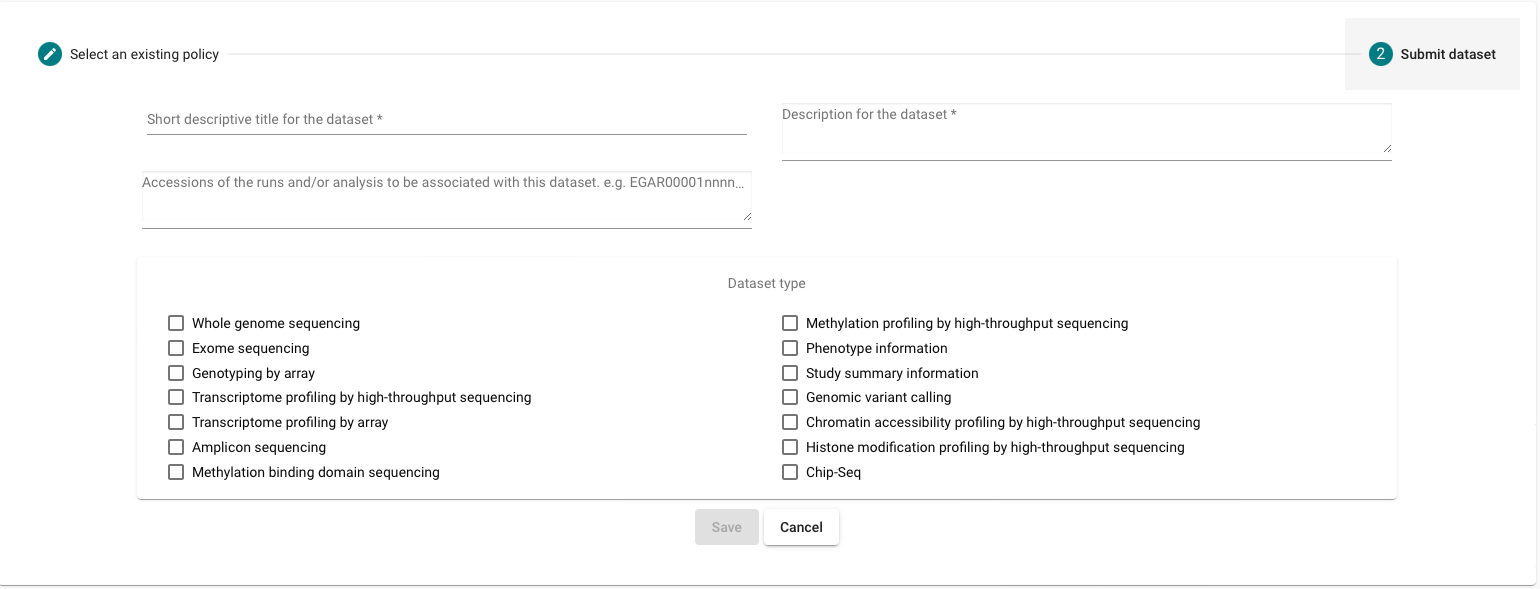
- After submitting your dataset you should contact the EGA Helpdesk to provide a release date for your dataset.
Datasets are automatically held (i.e. not released) unless they are affiliated to a study that has already been released.
Edit/update existing submission metadata
Go to the “Report” section of the object you would like to edit.
Locate the object and click on the arrow under action. An option menu will be displayed. Objects can be edited through their XML or with the WEBIN menu.
After an object has been edited, changes would be available on the website until the submission is released again. Please contact the EGA Helpdesk if you require further assistance.
